Code
library(tidyverse)
library(tidymodels)fit_resamples() and tune_grid() are tidymodels functions that we use in combination with objects generated from a resampling function (e.g., vfold_cv(), bootstraps(), etc from the rsample package) to get held-out performance estimates for our models.
fit_resamples() and tune_grid() use looping under the hood to accomplish their goals. fit_resamples() loops over the splits generated by our resampling function to get held-out performance estiamtes for a single model configuration.
tune_grid() also loops over these splits but includes an additional inner loop that loops over the values of our hyper-parameters in a hyper-parameter grid that we create.
In this appendix, we are going to re-create the functionality of fit_resamples() and tune_grid() using the map() function from the purrr package. This will help us understand how these functions work under the hood and give us a better understanding of how to use them. It will also give us an alternative way to do resampling if we need it.
Specifically, with respect to using resampling to get held-out performance estimates, we will
bootstraps() and vfold_cv()bootstraps() to both select a best model configuration and evaluate that best configuration with the variance benefits that are offered by nested cv relative to using a single held out test set.More generally, we will also gain a better understanding (and some code examples) of the use of list columns with iteration via map().
In this appendix, we will work through four separate examples that build in complexity
fit_resamples() computations by using map() to loop over the splits and fit a single model to each split. We use separate independent maps() for each stepmap() to loop that function over the splits.tune_grid() by using two nested map()s to loop over the splits and the hyper-parameter grid.map() Nested resampling includes both an outer and inner loop across different out and inner splits. There is also an innermost loop across values in a hyper-parameter grid (or any other grid that defines different model configurations)Lets start all of these examples by….
Loading libraries
library(tidyverse)
library(tidymodels)Creating a simple data set that has 300 observationns and two predictors (x1 and x2) and one outcome (y). The outcome is a linear combination of the two predictors with some noise added.
set.seed(123456)
n_obs <- 300
d <- tibble(x1 = rnorm(n_obs), x2 = rnorm(n_obs), y = 2*x1 + 3*x2 + rnorm(n_obs))Getting bootstrap resamples of d.
n_boots <- 50
resamples <- bootstraps(d, times = n_boots)bootstraps()
n_boots (in this case 50) rows. Each row is a bootstrap sample of the data. The columns are:splits - contains a bootstrap sample of the data that includes held-in raw data and OOB held-out raw data subsamplesid - name of the resampleresamples# Bootstrap sampling
# A tibble: 50 × 2
splits id
<list> <chr>
1 <split [300/115]> Bootstrap01
2 <split [300/108]> Bootstrap02
3 <split [300/108]> Bootstrap03
4 <split [300/107]> Bootstrap04
5 <split [300/112]> Bootstrap05
6 <split [300/102]> Bootstrap06
7 <split [300/112]> Bootstrap07
8 <split [300/114]> Bootstrap08
9 <split [300/115]> Bootstrap09
10 <split [300/117]> Bootstrap10
# ℹ 40 more rowsThis resamples object is a tibble (as typical in the tidy framework).
splits column in this tibble contain splits, a special object created by functions in the rsample package that hold resampled datasets.We can extract the contents of one of these cells using the base R [[]] notation
resamples$splits[[1]]<Analysis/Assess/Total>
<300/115/300>We will also need a recipe to create features from our raw data. Here is a simple recipe that indicate that y will be models on all the other columns (x1 and x2). We will not do any other feature engineering to keep this example simple.
rec <- recipe(y ~ ., data = d)We are now ready to start the first example
In this first example, we will combine map() with the use of list columns to save all the intermediate products that are produced when fitting and evaluating a single model configuration for a simple linear regression across many (in this case 50) held-out sets created by bootstrapping.
To do this, we will need a function to fit linear models to held-in training data. We can use the typical tidymodels code here.
fit_lm <- function(held_in) {
linear_reg() |>
set_engine("lm") |>
fit(y ~ ., data = held_in)
}Then we use map() and list columns to save the individual steps for evaluating the model in each split/resample. The following steps are done for EACH resample using map() or map2()
prep_recs column)new_data = NULL to get held-in features (in held_ins)new_data = assessment(split) to get held-out features (in held_outs)models)predictions)rmses)resamples_ex1 <- resamples |>
mutate(prep_recs = map(splits,
\(split) prep(rec, training = analysis(split))),
held_ins = map2(resamples$splits, prep_recs,
\(split, prep_rec) bake(prep_rec, new_data = NULL)),
held_outs = map2(resamples$splits, prep_recs,
\(split, prep_rec) bake(prep_rec,
new_data = assessment(split))),
models = map(held_ins,
\(held_in) fit_lm(held_in)),
predictions = map2(models, held_outs,
\(model, held_out) predict(model, held_out)$.pred),
rmses = map2_dbl(predictions, held_outs,
\(pred, held_out) rmse_vec(held_out$y, pred)))The pipline above creates a tibble with columns for each of the intermediate products and the rmse/error of the model in each resample.
map_dbl() to create the rmses column.resamples_ex1 |> glimpse()Rows: 50
Columns: 8
$ splits <list> [<boot_split[300 x 115 x 300 x 3]>], [<boot_split[300 x 1…
$ id <chr> "Bootstrap01", "Bootstrap02", "Bootstrap03", "Bootstrap04"…
$ prep_recs <list> [x1, x2, y, double, numeric, double, numeric, double, num…
$ held_ins <list> [<tbl_df[300 x 3]>], [<tbl_df[300 x 3]>], [<tbl_df[300 x …
$ held_outs <list> [<tbl_df[115 x 3]>], [<tbl_df[108 x 3]>], [<tbl_df[108 x …
$ models <list> [NULL, ~NULL, ~NULL, regression, FALSE, stats, formula, f…
$ predictions <list> <1.38587470, 2.84334628, -2.07435584, 1.24941793, -0.0569…
$ rmses <dbl> 0.9826968, 0.9380291, 1.0127994, 0.9929520, 0.9849202, 0.9…We can now look at rmses across the 50 bootstraps. For example, we can make a histogram using ggplot from the errors column in the fits tibble
resamples_ex1 |>
ggplot(aes(rmses)) +
geom_histogram(binwidth = 0.05)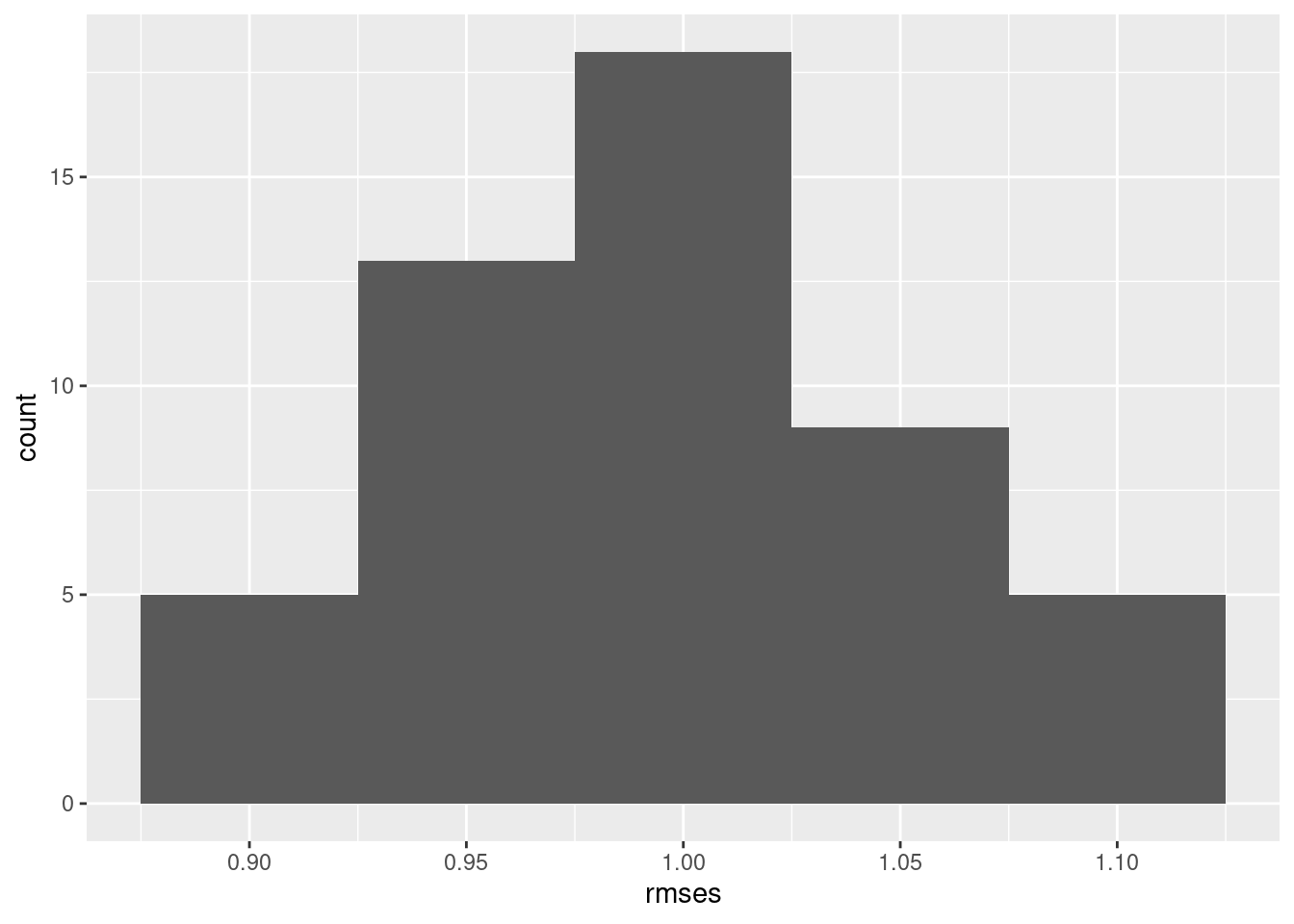
And we can summarize the min, max, mean, median and stdev of the error column in the fits tibble
resamples_ex1 |>
summarize(n = n(),
min = min(rmses),
max = max(rmses),
mean = mean(rmses),
median = median(rmses),
std_dev = sd(rmses))# A tibble: 1 × 6
n min max mean median std_dev
<int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 50 0.876 1.12 0.997 0.989 0.0557Easy peasy!
map() and list columns, which has many uses.fit_resamples() is doing.fit_resamples().
fit() function from keras to fit the model and then use predict() to get predictions. But you could still use the functions from the rsample package to get a resampled object (e.g., using bootstraps()) and you could use performance metrics (e.g., rmse_vec()) from the yardstick package.But remember, if you can (and in most instances, you can), it is easier to still using fit_resamples() to get your held-out performance estimates.
resamples_tidy_ex1 <-
linear_reg() |>
set_engine("lm") |>
fit_resamples(preprocessor = rec,
resamples = resamples,
metrics = metric_set(rmse))
resamples_tidy_ex1 |>
collect_metrics()# A tibble: 1 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 0.997 50 0.00788 Preprocessor1_Model1resamples_tidy_ex1 |>
collect_metrics(summarize = FALSE)# A tibble: 50 × 5
id .metric .estimator .estimate .config
<chr> <chr> <chr> <dbl> <chr>
1 Bootstrap01 rmse standard 0.983 Preprocessor1_Model1
2 Bootstrap02 rmse standard 0.938 Preprocessor1_Model1
3 Bootstrap03 rmse standard 1.01 Preprocessor1_Model1
4 Bootstrap04 rmse standard 0.993 Preprocessor1_Model1
5 Bootstrap05 rmse standard 0.985 Preprocessor1_Model1
6 Bootstrap06 rmse standard 0.947 Preprocessor1_Model1
7 Bootstrap07 rmse standard 1.00 Preprocessor1_Model1
8 Bootstrap08 rmse standard 1.06 Preprocessor1_Model1
9 Bootstrap09 rmse standard 1.05 Preprocessor1_Model1
10 Bootstrap10 rmse standard 0.981 Preprocessor1_Model1
# ℹ 40 more rowsIf we wanted to generate the held-out error using resampling but didnt need/want the intermediate products, we could wrap all the steps in one function and just map that single function.
Here is a function that takes a split and a recipe and returns the rmse of the model fit to the held-in data and evaluated on the held-out data. It does all the steps we did in the previous example but in one function. We will use this function to replace all the intermediate steps in the previous example.
fit_and_eval <- function(split, rec) {
# prep the recipe with held-in data
prep_rec <- prep(rec, training = analysis(split))
# bake the recipe using new_data = NULL to pull out the held-in features
held_in <- bake(prep_rec, new_data = NULL)
# bake the recipe using new_data = assessment(split) to get held-out features
held_out <- bake(prep_rec, new_data = assessment(split))
# fit the model using the held-in features
model <-
linear_reg() |>
set_engine("lm") |>
fit(y ~ ., data = held_in)
# get predictions using the model with the held-out features
pred <- predict(model, held_out)$.pred
# calculate the accuracy of the model
rmse_vec(held_out$y, pred)
}Now map this function over the splits to get a vector of rmse. Same results, but not saving intermediate steps by using one function.
resamples_ex2 <- resamples |>
mutate(errors = map_dbl(splits, \(split) fit_and_eval(split, rec)))Here is what the resamples_ex2 tibble now looks like
resamples_ex2# Bootstrap sampling
# A tibble: 50 × 3
splits id errors
<list> <chr> <dbl>
1 <split [300/115]> Bootstrap01 0.983
2 <split [300/108]> Bootstrap02 0.938
3 <split [300/108]> Bootstrap03 1.01
4 <split [300/107]> Bootstrap04 0.993
5 <split [300/112]> Bootstrap05 0.985
6 <split [300/102]> Bootstrap06 0.947
7 <split [300/112]> Bootstrap07 1.00
8 <split [300/114]> Bootstrap08 1.06
9 <split [300/115]> Bootstrap09 1.05
10 <split [300/117]> Bootstrap10 0.981
# ℹ 40 more rowsAnd here we demo getting overall held-out performance details across the 50 bootstraps.
resamples_ex2 |>
summarize(n = n(),
min = min(errors),
max = max(errors),
mean = mean(errors),
median = median(errors),
std_dev = sd(errors))# A tibble: 1 × 6
n min max mean median std_dev
<int> <dbl> <dbl> <dbl> <dbl> <dbl>
1 50 0.876 1.12 0.997 0.989 0.0557Now we can make this a bit more complicated by adding a grid of hyperparameters to tune.
tune_grid() but we can do it again with two loops using map().map() to loop over the resamples (as we did in the last two examples) and an inner map() to loop over the values of kLets start by setting up a grid of values of k to tune over.
grid_k = tibble(neighbors = c(3, 6, 9, 12, 15, 18))We will use a single function to repeatedly fit and eval over our grid of parameters.
eval_grid <- function(split, rec, grid_k) {
# get held-in and held-out features for split
# we calculate features inside the function where we fit all models across the grid
# because we want to make sure we only need to prep and bake the recipe once
# per split. Otherwise, we would waste a lot of computational time.
prep_rec <- prep(rec, training = analysis(split))
held_in <- bake(prep_rec, new_data = NULL)
held_out <- bake(prep_rec, new_data = assessment(split))
# function to fit and eval model for a specific k using held-in/held-out
# for this split
fit_eval <- function(k, held_in, held_out) {
model <-
nearest_neighbor(neighbors = k) |>
set_engine("kknn") |>
set_mode("regression") |>
fit(y ~ ., data = held_in)
pred <- predict(model, held_out)$.pred
# lets put k and rmse in a tibble and return that for each split
tibble(k = k,
rmse = rmse_vec(held_out$y, pred))
}
# loop through grid_k and fit/eval model for each k
# this is the inner loop from tune_grid()
# use list_rbind() to bind the separate rows for each tibble into one larger tibble
grid_k$neighbors |>
map(\(k) fit_eval(k, held_in, held_out)) |>
list_rbind()
}Now we map this function over the 50 bootstrap splits.
tune_grid().eval_grid() will return a tibble with rows for each value of k. We will save one tibble for each split in a list column called `rmses``.resamples_ex3 <- resamples |>
mutate(rmses = map(splits,
\(split) eval_grid(split, rec, grid_k)))The rmses column contains the rmse for each value of k for each split/resample in a tibble. Each tibble has 6 rows (one for each k) and 2 columns (k and rmse).
resamples_ex3# Bootstrap sampling
# A tibble: 50 × 3
splits id rmses
<list> <chr> <list>
1 <split [300/115]> Bootstrap01 <tibble [6 × 2]>
2 <split [300/108]> Bootstrap02 <tibble [6 × 2]>
3 <split [300/108]> Bootstrap03 <tibble [6 × 2]>
4 <split [300/107]> Bootstrap04 <tibble [6 × 2]>
5 <split [300/112]> Bootstrap05 <tibble [6 × 2]>
6 <split [300/102]> Bootstrap06 <tibble [6 × 2]>
7 <split [300/112]> Bootstrap07 <tibble [6 × 2]>
8 <split [300/114]> Bootstrap08 <tibble [6 × 2]>
9 <split [300/115]> Bootstrap09 <tibble [6 × 2]>
10 <split [300/117]> Bootstrap10 <tibble [6 × 2]>
# ℹ 40 more rowsLets take a look at one of these tibbles to make this structure clearer. The same tibble format is saved for all fifty bootstrap splits (with different values for rmse of course!)
resamples_ex3$rmses[[1]]# A tibble: 6 × 2
k rmse
<dbl> <dbl>
1 3 1.51
2 6 1.47
3 9 1.47
4 12 1.52
5 15 1.56
6 18 1.61We can unnest() the rmses column to get a tibble with one row for each k value in each resample.
unnest() is used frequently when a list column contains tibbles and you want to combine those tibbles into more traditional tibble without the list column.resamples_ex3 |>
unnest(rmses) |>
select(-splits) |>
print(n = 30)# A tibble: 300 × 3
id k rmse
<chr> <dbl> <dbl>
1 Bootstrap01 3 1.51
2 Bootstrap01 6 1.47
3 Bootstrap01 9 1.47
4 Bootstrap01 12 1.52
5 Bootstrap01 15 1.56
6 Bootstrap01 18 1.61
7 Bootstrap02 3 1.31
8 Bootstrap02 6 1.25
9 Bootstrap02 9 1.25
10 Bootstrap02 12 1.27
11 Bootstrap02 15 1.29
12 Bootstrap02 18 1.30
13 Bootstrap03 3 1.48
14 Bootstrap03 6 1.46
15 Bootstrap03 9 1.48
16 Bootstrap03 12 1.50
17 Bootstrap03 15 1.52
18 Bootstrap03 18 1.54
19 Bootstrap04 3 1.41
20 Bootstrap04 6 1.34
21 Bootstrap04 9 1.31
22 Bootstrap04 12 1.32
23 Bootstrap04 15 1.35
24 Bootstrap04 18 1.38
25 Bootstrap05 3 1.51
26 Bootstrap05 6 1.43
27 Bootstrap05 9 1.41
28 Bootstrap05 12 1.42
29 Bootstrap05 15 1.44
30 Bootstrap05 18 1.44
# ℹ 270 more rowsWe can pipe this unnested tibble into a group_by() and summarize() to get the median (or mean) across resamples and then arrange to find the best k
resamples_ex3 |>
unnest(rmses) |>
select(-splits) |>
group_by(k) |>
summarize(n = n(),
mean_rmse = mean(rmse)) |>
arrange(mean_rmse)# A tibble: 6 × 3
k n mean_rmse
<dbl> <int> <dbl>
1 9 50 1.34
2 12 50 1.35
3 6 50 1.36
4 15 50 1.36
5 18 50 1.38
6 3 50 1.42The example makes it clear that using resampling to tune a grid of hyperparameters is just a matter of looping over the resamples in an outer loop and looping over a grid of hyperparameters in an inner loop.
You might use this approach
But of course, when you can use it, using tune_grid() is easier because it takes care of the nested looping internally.
resamples_tidy_ex3 <-
nearest_neighbor(neighbors = tune()) |>
set_engine("kknn") |>
set_mode("regression") |>
tune_grid(preprocessor = rec,
resamples = resamples,
grid = grid_k,
metrics = metric_set(rmse))
resamples_tidy_ex3 |>
collect_metrics() |>
arrange(mean)# A tibble: 6 × 7
neighbors .metric .estimator mean n std_err .config
<dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
1 9 rmse standard 1.34 50 0.0184 Preprocessor1_Model3
2 12 rmse standard 1.35 50 0.0202 Preprocessor1_Model4
3 6 rmse standard 1.36 50 0.0165 Preprocessor1_Model2
4 15 rmse standard 1.36 50 0.0216 Preprocessor1_Model5
5 18 rmse standard 1.38 50 0.0225 Preprocessor1_Model6
6 3 rmse standard 1.42 50 0.0161 Preprocessor1_Model1Now lets do the most complicated version of this. Nested resampling involves looping over outer splits where the held out data are test sets used to evaluate best model configurations for each outer split and the inner loop makes validation sets that are used to select the best model configuration for each outer fold. However, if we are tuning a grid of hyperparameters, there is even a further nested loop inside the inner resampling loop to get performance metrics for each value of the hyperparameter in the validation sets.
rsample supports creating a nested resampling object.
fit_resamples() or tune_grid() support nested resampling. So we will always need to use nested map() to do this ourselves.First, lets make the nested resampling object and explore it a bit
resamples_nested <- d |>
nested_cv(outside = vfold_cv(v = 5), inside = bootstraps(times = 10))Lets take a look at it.
resamples_nested# Nested resampling:
# outer: 5-fold cross-validation
# inner: Bootstrap sampling
# A tibble: 5 × 3
splits id inner_resamples
<list> <chr> <list>
1 <split [240/60]> Fold1 <boot [10 × 2]>
2 <split [240/60]> Fold2 <boot [10 × 2]>
3 <split [240/60]> Fold3 <boot [10 × 2]>
4 <split [240/60]> Fold4 <boot [10 × 2]>
5 <split [240/60]> Fold5 <boot [10 × 2]>Lets look a little more carefully at this structure because its critical to understanding nested cv.
resamples_nested$splits[[1]]<Analysis/Assess/Total>
<240/60/300># I prefer to work with this resampled objects using base R notation
# however you could do the same using piped tidy code
# resamples_nested |>
# slice(1) |>
# pull(splits)Now look below at the inner resamples for the first outer k-fold split.
resamples_nested$inner_resamples[[1]]# Bootstrap sampling
# A tibble: 10 × 2
splits id
<list> <chr>
1 <split [240/87]> Bootstrap01
2 <split [240/86]> Bootstrap02
3 <split [240/80]> Bootstrap03
4 <split [240/76]> Bootstrap04
5 <split [240/89]> Bootstrap05
6 <split [240/93]> Bootstrap06
7 <split [240/90]> Bootstrap07
8 <split [240/92]> Bootstrap08
9 <split [240/93]> Bootstrap09
10 <split [240/87]> Bootstrap10Now that we have a resamples object to use for nested cv, lets get some functions together to calculate rmses for inner validation sets and outer test sets
We will need a function again that takes a split, uses the recipe to get held-in/held-out features, and then fits and evaluates a model for each value of k in the grid. This is the exact same function as we used earlier (because we are using knn again). We include it below again for your review.
eval_grid <- function(split, rec, grid_k) {
# get held-in and held-out features for split
prep_rec <- prep(rec, training = analysis(split))
held_in <- bake(prep_rec, new_data = NULL)
held_out <- bake(prep_rec, new_data = assessment(split))
# function fit fit and eval model for a specific lambda and resample
fit_eval <- function(k, held_in, held_out) {
model <-
nearest_neighbor(neighbors = k) |>
set_engine("kknn") |>
set_mode("regression") |>
fit(y ~ ., data = held_in)
pred <- predict(model, held_out)$.pred
# lets put k and rmse in a tibble and return that for each split
tibble(k = k,
rmse = rmse_vec(held_out$y, pred))
}
# loop through grid_k and fit/eval model for each k
# this is the inner loop
# use list_rbind() to bind the separate rows for each tibble into one larger tibble
grid_k$neighbors |>
map(\(k) fit_eval(k, held_in, held_out)) |>
list_rbind()
}To loop eval_grid over the 10 bootstraps in the nested cv inner loop, we will need another simple function that applies our eval_grid() over each bootstrap within a set of 10 and binds the results into a single tibble.
bind_bootstraps <- function(bootstraps, rec, grid_k) {
bootstraps$splits |>
map(\(bootstrap_split) eval_grid(bootstrap_split, rec, grid_k)) |>
list_rbind()
}Now we are ready to get performance metrics for each k in grid_k for each of the inner 10 bootstraps in each of the five outer k-folds splits. Remember that bind_bootstraps() just calls eval_grid() for each of the 10 bootstraps in the inner loop and binds the results.
resamples_nested_ex4 <- resamples_nested |>
mutate(inner_rmses = map(inner_resamples,
\(inner_resample) bind_bootstraps(inner_resample,
rec,
grid_k)))Lets look at what we just added to our resamples object
resamples_nested_ex4 # Nested resampling:
# outer: 5-fold cross-validation
# inner: Bootstrap sampling
# A tibble: 5 × 4
splits id inner_resamples inner_rmses
<list> <chr> <list> <list>
1 <split [240/60]> Fold1 <boot [10 × 2]> <tibble [60 × 2]>
2 <split [240/60]> Fold2 <boot [10 × 2]> <tibble [60 × 2]>
3 <split [240/60]> Fold3 <boot [10 × 2]> <tibble [60 × 2]>
4 <split [240/60]> Fold4 <boot [10 × 2]> <tibble [60 × 2]>
5 <split [240/60]> Fold5 <boot [10 × 2]> <tibble [60 × 2]>If we look at one of them, we see
resamples_nested_ex4$inner_rmses[[1]]# A tibble: 60 × 2
k rmse
<dbl> <dbl>
1 3 1.52
2 6 1.54
3 9 1.58
4 12 1.62
5 15 1.65
6 18 1.68
7 3 1.77
8 6 1.70
9 9 1.69
10 12 1.70
# ℹ 50 more rowsWe now need to calculate the best k for each of the sets of 10 boostraps (associated with one outer k-fold split). To do this, we average the 10 rmses together for each value of k and then choose the k with the lowest rmse. We will write a function to do this because we can then map that function over each of the sets of 10 bootstraps associated with each outer split.
get_best_k <- function(rmses) {
rmses |>
group_by(k) |>
summarize(mean_rmse = mean(rmse)) |>
arrange(mean_rmse) |>
slice(1) |>
pull(k)
}And then map it across the sets of rmses for each outer k-fold splilt.
resamples_nested_ex4 <- resamples_nested_ex4 |>
mutate(best_k = map_dbl(inner_rmses, \(rmses) get_best_k(rmses)))Now we have determined the best value of k for each of the 5 outer folds.
resamples_nested_ex4# Nested resampling:
# outer: 5-fold cross-validation
# inner: Bootstrap sampling
# A tibble: 5 × 5
splits id inner_resamples inner_rmses best_k
<list> <chr> <list> <list> <dbl>
1 <split [240/60]> Fold1 <boot [10 × 2]> <tibble [60 × 2]> 6
2 <split [240/60]> Fold2 <boot [10 × 2]> <tibble [60 × 2]> 12
3 <split [240/60]> Fold3 <boot [10 × 2]> <tibble [60 × 2]> 12
4 <split [240/60]> Fold4 <boot [10 × 2]> <tibble [60 × 2]> 6
5 <split [240/60]> Fold5 <boot [10 × 2]> <tibble [60 × 2]> 9We can now move up to the outer k-fold splits. We will use the best k for each outer fold to fit a model using the held-in data from the outer fold and then evaluate that model using the held-out test data. Those test data were NEVER used before so the will allow us to see how that model with a best value of k (selected by bootstrap resampling) performs in new data
We can reuse the eval_grid() function we used before to fit and evaluate the model using the held-in data and the held-out test data. But now we just use one value of k, the one that was best for each fold
resamples_nested_ex4 <- resamples_nested_ex4 |>
mutate(test_rmse = map2(splits, best_k,
\(split, k) eval_grid(split, rec, tibble(neighbors = k))))We can pull the one row tibbles out of this final column and bind them together to get a tibble with one row for each outer fold. We have retained the best value of k for each fold for our information (these don’t need to be the same for each k-fold split).
resamples_nested_ex4$test_rmse |>
list_rbind() |>
mutate(outer_fold = 1:length(resamples_nested_ex4$test_rmse)) |>
relocate(outer_fold)# A tibble: 5 × 3
outer_fold k rmse
<int> <dbl> <dbl>
1 1 6 1.23
2 2 12 1.58
3 3 12 1.28
4 4 6 1.21
5 5 9 1.06If we want to know the true best k to use when we train a final model to implement using all our data, we need select it by using our inner reasampling method (10 boostraps) with all our data.
We could then train our final implementation model by fitting a model to all our data using this value of k.
All of these examples were done without the use of parallel processing. fit_resamples() and tune_grid() can use a parallel backend if you set it up.
map() in parallel using future_map() from the furrr package.fit_resamples() examples, there is only one map() or several independent ones. None are nested. Switch them all to future_map().tune_grid() example, there are two nested maps(). In most instances, you will want to switch the outer loop map() to future_map() and leave the inner map() as is.map()s. In most instances, you will want to switch that outermost map() (across outer splits) to future_map() and leave the inner two map()s as is. - See our post on setting up parallel backends for more info on how to do that